Easily Find the Revolution Sample Prep Kit Best Suited to Your Assay's Analyte, Sample Type, and Input Sample Volume. All Kits are for Use on the Revolution Plus Instrument.
The Revolution Plus Platform
Smart, easy-to-use semi-automated extraction using nRichDX's Revolution kits for cfDNA, cfTNA/cfRNA, and CTCs.
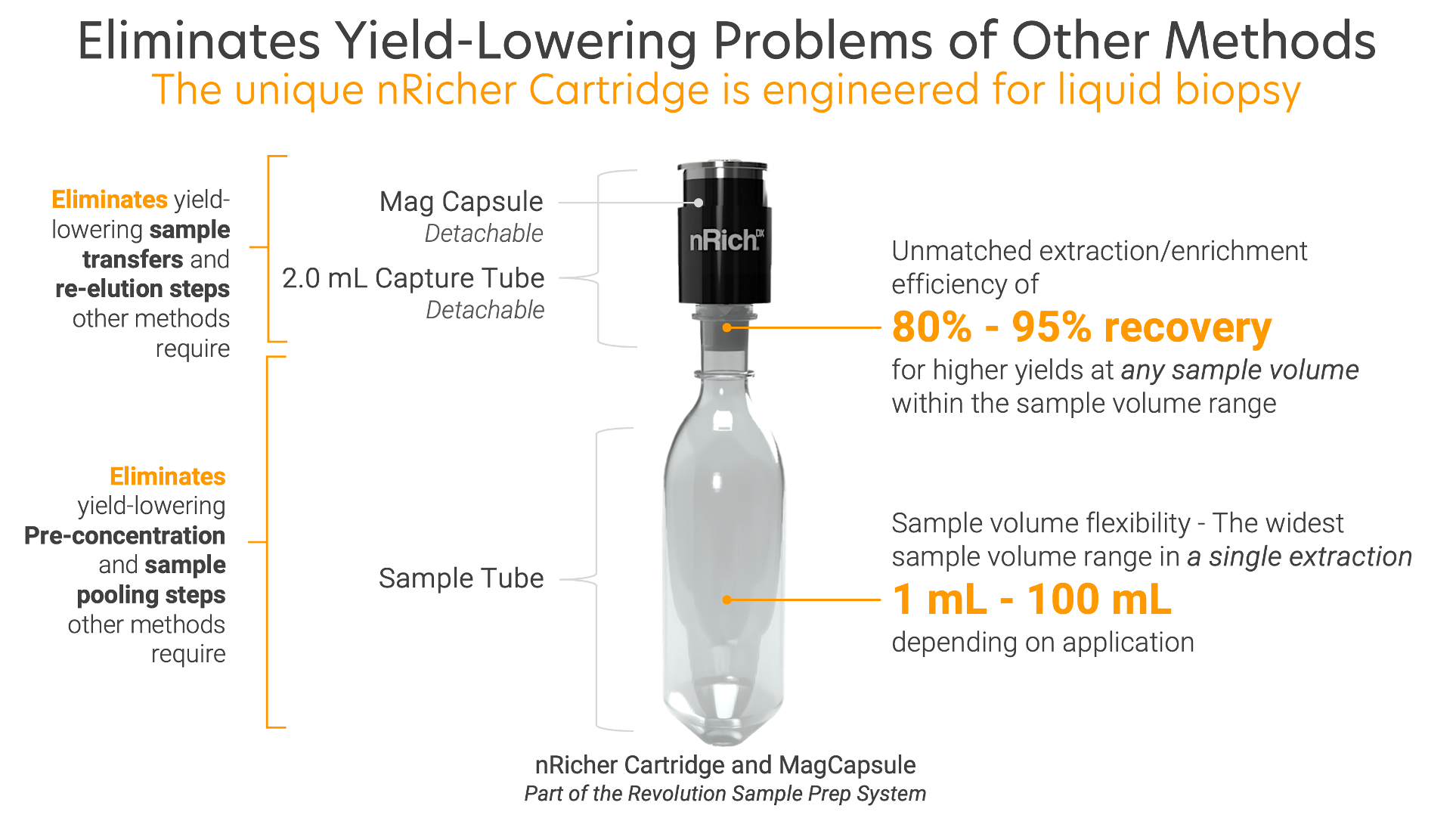
The Cartridge Makes the Difference
The unique nRicher Cartridge maximizes yield and purity by eliminating problems found in other methods. Only from nRichDX.
Automated Solutions with Hamilton
As a Hamilton Trusted Partner, nRichDX's Revolution System and the Hamilton Microlab STAR liquid handler combine to create fully scalable automated liquid biopsy sample solutions.
SABCS 2025 Poster - Detecting Breast Cancer Mutations in 50 mL Urine
Considering urine as a liquid biopsy biofluid for detection of cancer markers? Check out nRichDX's recent poster at the San Antonio Breast Cancer Symposium. Presented in collaboration with Anchor Molecular.
See Our AMP 2025 Posters
nRichDX and collaborators presented four posters at the AMP 2025 Meeting & Expo in Boston. See them in detail on our Reference Materials webpage.
What Users Say
"In our work with both cancer and neurological disease samples, the nRichDX platform has consistently given us high-quality, high-yield cfDNA. The workflow is straightforward and reliable, and it has genuinely improved the sensitivity and confidence of our liquid biopsy studies."
On-Demand Webinar Video Now Available
Title: "Current Perspectives on RNA Liquid Biopsy: Impact of RNA Extraction on Assay Performance"
Date/Time: Held Thursday, October 16th at 9am PST/Noon EST/6pm CET
Investigating Cell-Free RNA?
nRichDX's Revolution cfTNA Max 20 and Max 50 Kits extract cell-free circulating Total Nucleic Acid, including cfRNA and ctRNA.
Johns Hopkins University Lab Publication Features Revolution System
Full-Length Clonal Immunoglobulin Rearrangements in cfDNA: Improved Recovery and Sequencing by Xian, R. et. al
Dartmouth Lab Validates Revolution System
Standardized Workflow and Analytical Validation of Cell-Free DNA Extraction for Liquid Biopsy Using a Magnetic Bead-Based Cartridge System by Sathyanarayana, S. et al.
RNA Extraction Comparison
ASHG 2025 poster (Genewiz) concludes, "The nRichDX Revolution Plus System yielded more and higher-quality cfRNA, with measurable RIN values where other kits failed. This improved input enabled more sensitive detection of ALS-associated cryptic introns."
Not Sure Which Revolution Kit to Use?
Use the Revolution Kit Selector or view a summary table of Revolution Sample Prep Kits currently available on the Revolution Plus Sample Prep System.
The Overlooked Role of Sample Prep in Limiting Liquid Biopsy Sensitivity
Discover why spin columns fall short for rare liquid biopsy analyte extraction, particularly when scaling input volumes to more than 5 mL.
The Revolution Sample Prep Platform
For your liquid biopsy requirements today... and tomorrow
The Simple Math
Analyte yield is a function of input sample volume and recovery rate
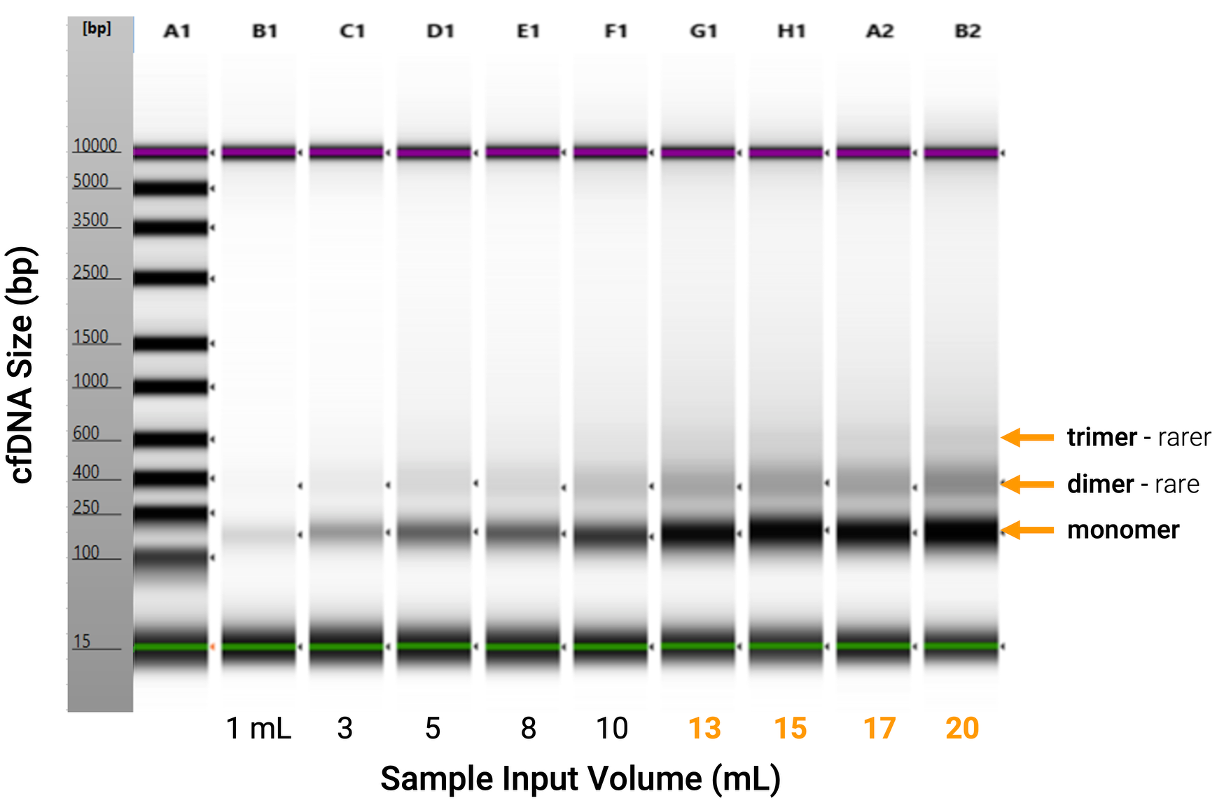
As sample volume increases, analyte yield increases. Note the increasing intensity of rarer species of cfDNA indicated by the dimer and trimer forms with increasing sample volume. Competing methods are often limited to 10 mL or less of input sample volume per extraction.
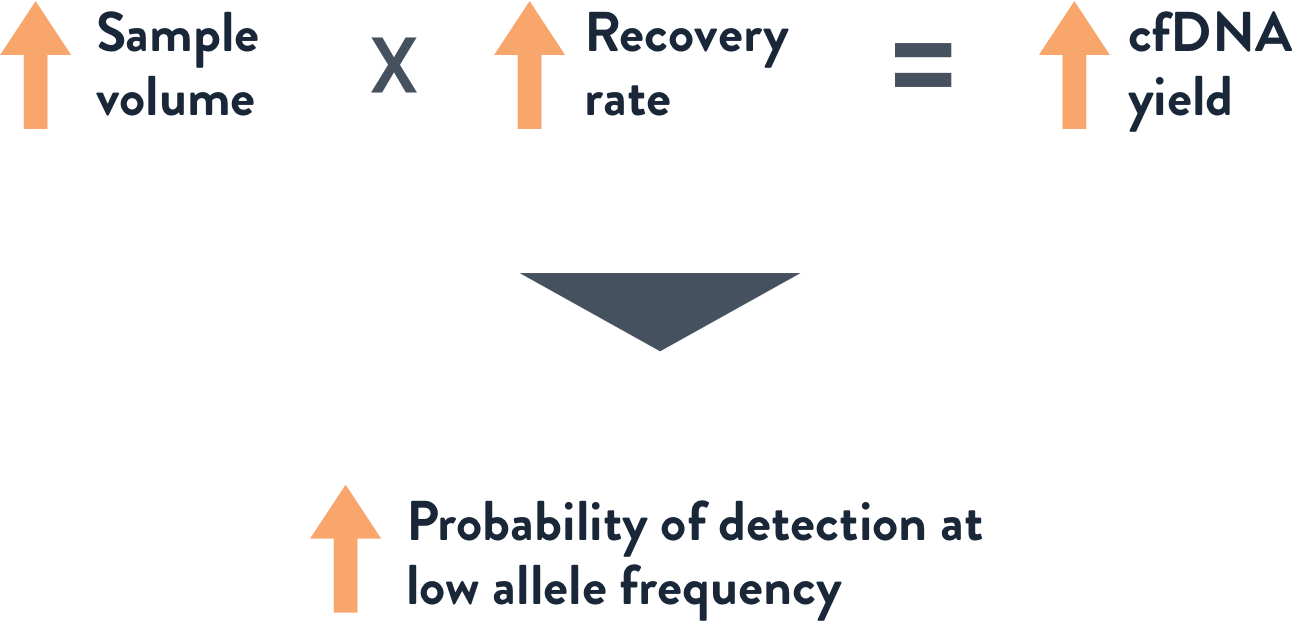
Probability of detection of rare alleles at a low VAF of 0.1% or even 0.01% depends on a sufficiently high cfDNA input into the detection assay. Optimal input is a simple function of sample volume and recovery rate. Competing sample prep systems are lacking in both sample volume capacity and recovery rate, and therefore don’t produce sufficient yield to ensure a high probability of allele detection. This is a leading cause of Quantity Not Sufficient (QNS) errors in liquid biopsy assays.
Liquid Biopsy's Sample Prep Problem
Existing Methods: Inherent Limitations
Other liquid biopsy extraction methods use yield-lowering procedures
- Sample Transfers - moving the sample from plasticware to plasticware during the extraction protocol, leaving small amounts of material behind with each sample transfer - including targeted rare analytes
- Pre-extraction Sample Volume Concentration - Reducing the input sample volume to accommodate inherent volume limitations of the method
- Re-elution of the extraction eluate - attempting to increase yield of analyte by passing the eluted sample over the extraction matrix repeatedly to boost yield of rare analytes
- Pooling - extracting the same sample using multiple separate extractions and combining the eluates, which may require subsequent eluate volume concentration
- Limited Input Sample Volumes - other methods usually have inherent maximum input sample volumes limited to less than or equal to 5 mL - 10 mL which may be insufficient for early detection of rare analytes*
*Other methods, including automated methods, are inherently sample-volume limited and typically must use one or more of the above yield-lowering schemes to extract from volumes greater than 5 mL - 10 mL. These methods were not specifically engineered for extraction of rare analytes such as ctDNA and ctRNA from larger volumes which may lead to lower yields. Lower yields of analytes in the assay lowers assay sensitivity and limit of detection (LoD) and may result in Quantity Not Sufficient (QNS) errors.
Easy Workflow - Automation for the nRicher Cartridge
The nRichDX Revolution Plus Instrument
Extraction of up to 12 samples per run in a semi-automated workflow, exclusively with Revolution Kits

Revolution Kit Selector

- cf/ctDNA
- cf/ctRNA
- cf/ctTNA (Total Nucleic Acid)
- CTCs (Circulating Tumor Cells)
- Whole Blood
- Plasma
- Urine
- CSF
- Peritoneal Fluid
- 1 mL – 20 mL
- >20 mL – 50 mL
- >50 mL





Revolution cfTNA Max 20 Kit

Revolution cfTNA Max 20 Reagent Kit

Revolution cfTNA Max 50 Kit

Revolution cfTNA Max 50 Reagent Kit

Revolution CTC Enrichment Kit (Epithelial Origin)

Revolution CTC Reagent Kit
No Product Found. For more information, please contact your Salesperson or email nRichDX at info@nrichdx.com
Note: your price may be different than the list prices shown. To inquire about, or request a quote, please click here, or email to info@nrichdx.com.
Interested in a different sample type? Please email to info@nrichdx.com.